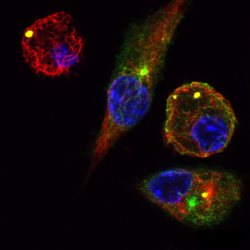
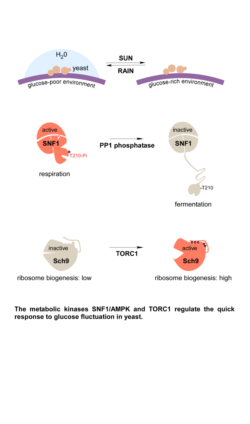
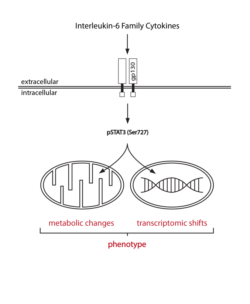
Metabolites and the according metabolic pathways are not merely providing energy and building blocks to sustain cellular and organismal function but can carry and transmit information, affecting other components of biological systems. This is obvious for situations where a drop in concentration of certain metabolites has to be detected to mount a proper response, i.e. increasing uptake or generation, reducing consumption or promoting recycling of metabolites. It becomes increasingly clear that the role of metabolism in signalling goes far beyond these basic mechanisms and that there are multifaceted connection points between metabolism and other mechanism of signalling. Freiburg Metabolism researchers study the role of specific metabolites and metabolic pathways in particular cell types and their responses, e.g. in the context of development and immunology. The community is increasingly interested in manipulating metabolism through control of function approaches.
Bertram Bengsch 
Faculty of Medicine and Medical Center – Department of Medicine II (Gastroenterology, Hepatology, Endocrinology and Infectious Disease)
RESEARCH: Immunometabolism, Metabolic Signalling, Mitochondria, Single-cell Immune Profiling, Mass Cytometry, Imaging Mass Cytometry, Adaptive Immunity, T cells, T cell exhaustion, Signal Transduction, Inflammatory Diseases, Infectious Diseases, Liver Diseases, Cancer Immunology and Immunotherapy
METHODS: Metabolism-directed flow and mass cytometry, imaging Pharmacologic and Genetic Manipulation of Metabolism, Extracellular Metabolic Flux Analysis, Untargeted Metabolomics by MS and NMR (outsourced and via collaborations)
TECHNOLOGIES: Mass cytometry, Imaging Mass cytometry, Hypoxia chambers, Flow cytometry
CIBSS, SFB/TRR 167, SFB 1160, SFB 1479, DKTK
Olaf Groß 
Faculty of Medicine and Medical Center – Institute of Neuropathology
RESEARCH: Immunometabolism, Cancer Metabolism, Metabolic Signalling, Mitochondria, Innate Immune Sensing, Macrophage Signal Transduction and Cell Biology, Cell Death, Inflammatory, Infectious, Cardiovascular and Nervous System Diseases
METHODS: Pharmacologic and Genetic Manipulation of Metabolism, Extracellular Metabolic Flux Analysis, Small Molecule Screening, Untargeted Metabolomics by MS and NMR (outsourced and via collaborations)
TECHNOLOGIES: Seahorse XFe96 Flux Analyzer (Agilent), Hypoxic Workstations i2 and H35 (Whitley), Multidrop Reagent Dispenser (Thermo)
Ralf Baumeister 
Faculty of Biology and Medicine – Institute of Biology 3, Bioinformatics and Molecular Genetics, ZBMZ
RESEARCH: Signaling mechanisms responding to stress and nutrients, ageing and age-related disesases, molecular genetics and biochemistry, C. elegans models of diseases. mTORC1 and mTORC2, SGK-1, signaling mechanisms in tumor biology, programmed cell death, autophagy
METHODS: Genetic dissection of signaling pathways, C. elegans models of human diseases, large scale RNAi screenings, protein interaction studies, DNA and RNA binding, stress biology
TECHNOLOGIES: genome-wide RNAi screenings, mutant screenings, hypoxia chamber, confocal microscopy, C. elegans robotics
Clemens Kreutz 
Faculty of Medicine and Medical Center – Institute of Medical Biometry and Statistics (IMBI), Methods of Systems Biomedicine (MSB)
RESEARCH: Mathematical Modelling, Metabolic Signaling Networks, Link to Microbiome, Link to Proteome, Methods for Single Cell Analyses
METHODS: Mathematical Models, Benchmarking and Optimal Selection of Computational Methods, Development of Novel Analysis Approaches
TECHNOLOGIES: High Performance Computing
Melanie Börries 
Faculty of Medicine and Medical Center – Institute of Medical Bioinformatics and Systems Medicine
RESEARCH: Multi-Omics data analysis, cancer metabolism, (genetic) analyses of metabolic signaling, identification of disease biomarkers and potential therapeutic options, pipeline development for high-throughput data for clinical research and applications (e.g. Molecular Tumor Board)
METHODS: machine learning methods, development and application of standardized workflows for the analysis, integration and visualization of omics data, single-cell analysis, creating user-friendly databases
TECHNOLOGIES: High-performance computer cluster, 10x Genomics
Claudia Jessen-Trefzer 
Faculty of Chemistry and Pharmacy – Institute of Organic Chemistry
RESEARCH: Infectious Diseases, Mycobacteria, Mycobacterial Metabolite Transporters, Metabolic Signalling, Signal Transduction, Extracellular Nanocompartments, Synthetic Biology, Mycobacterial Membranes, Antibiotics, Drug or Photosensitizer-Metabolite Conjugates and Organometallic Complexes
METHODS: Genetic Manipulation of Metabolism, Small Molecule Screening, Targeted Metabolomics by MS, Protein Isolation and Characterization, Proteomics (in collaboration), Molecular Biology, Microbiology, Organic Synthesis
TECHNOLOGIES: LED array 96-well plate (Teleopt), ChemStudio (Analytics Jena)
Miriam Schmidts 
Faculty of Medicine and Medical Center – Center for Pediatrics and Adolescent Medicine, Pediatric genetics Division
RESEARCH: Human Genetics, Developmental Biology, Signal Transduction, Cell Biology, Ciliopathies, Kidney Diseases, Skeletal defects, Neurodevelopmental Diseases
METHODS: NGS, Gene editing, Transcriptomics, Proteomics (collaboration with Oliver Schilling. Pathology, University Hospital Freiburg), Metabolomics (collaboration with prof. Spiekerkötter), EM (in external collaboration)
TECHNOLOGIES: CRISPR/Cas gene editing vectors, NGS
Andreas Bechthold 
Faculty of Chemistry and Pharmacy – Institute of Pharmaceutical Sciences, Pharmaceutical Biology and Biotechnology
RESEARCH: Metabolism, metabolism signaling, Natural products, antibiotics, biosynthesis of natural products, regulation, cyclic di-GMP, cofactor dependent enzymes, transport of natural products, oxygenases, glycoyltrasnferases, microorganisms in insects
METHODS: Genetic Manipulation of Metabolism, Small Molecule Structure elucidation, Characterization of Natural Products by MS and NMR, Protein structure elucidation, Cryo-EM
TECHNOLOGIES: Triple-Stage-Quadrupol-Massenspektrometer 40-500 + UnliMate 3000 Binäres HPLC-System + DAD (Thermo)
Chris Meisinger 
Medical Faculty – Institute of Biochemistry and Molecular Biology
RESEARCH: Dynamic Adaptation of the Mitochondrial Proteome upon Stress and Metabolic Switches, Metabolic Signalling, Mitochondria, Signal Transduction, Cell Biology, Nervous System Diseases, Oncogenic Signalling
METHODS: Mitochondrial and Submitochondrial Proteomics and Phosphoproteomics, Global Kinome Profiling for Mitochondrial Signalling Pathways, Import and Assembly of Mitochondrial Proteins, Extracellular Metabolic Flux Analysis
TECHNOLOGIES: Seahorse XFe96 Flux Analyzer (Agilent)
Katrin Kierdorf 
Faculty of Medicine and Medical Center – Institute of Neuropathology
RESEARCH: Developmental Immunology, Macrophage Development, Macrophage Function, Immunometabolism, Macrophage Metabolism, Metabolic Signalling
METHODS: Ex vivo cell culture platform to study immunometabolism in resident tissue macrophages, tissue normoxic cell cultures, transgenic Drosophila to study conserved immunometabolic signalling
CIBSS, SFB/TRR 167, CRC 1479
Tanja Vogel 
Faculty of Medicine – Institute of Anatomy and Cell Biology, Deptartment Molecular Embryology
RESEARCH: Epigenetic Modification impacting Metabolism, Developmental Processes associated with Metabolic Programs, Nervous System Diseases, Cell Biology, Signal Transduction, Metabolic States driving Stem Cell Specification
METHODS: iPSC, mESC, high throughput data generation and multi-omics analysis, single cell transcriptomics and imaging
TECHNOLOGIES: iPSC and mESC neuronal differentiation; CRISPR/Cas9; RNA- (bulk and single cell), ChIP- and ATAC-seq
Simone Hettmer 
Faculty of Medicine and Medical Center – Center for Pediatrics, Department of Pediatric Hematology and Oncology
RESEARCH: Cancer Metabolism, Metabolic Signalling, Signal transduction, Amino Acids, Developmental Biology, Signal Transduction, Cell Biology, Sarcomas, Oncogenic regulation of tumor metabolism, Antimetabolites as anti-cancer therapeutic agents
METHODS: Pharmacologic and Genetic Manipulation of Metabolism and Signaling in tumor cells, Extracellular Metabolic Flux Analysis, Small Molecule Screening, Untargeted Metabolomics by MS and NMR (outsourced and via collaborations)
TECHNOLOGIES: Mouse and tissue culture models of sarcoma
Dennis Wolf 
Faculty of Medicine and Medical Center – Cardiology and Angiology I, Vascular Immunology Laboratory
RESEARCH: Acquired Metabolic Disease, Metabolic Signalling, Immunometabolism, Adaptive Immunity, T cells, Signal Transduction, Cell Biology, Inflammatory Diseases, Cardiovascular Diseases, Apolipoproteins, Autoimmunity
METHODS: Flow Cytometry, CyTOF, single cell RNA-sequencing, clinical studies, Pharmacologic and genetic manipulation of metabolism, Transcriptomics
TECHNOLOGY: Comprehensive Lab Animal Monitoring System (CLAMS)
Manfred Jung 
Faculty of Chemistry and Pharmacy – Institute of Pharmaceutical Sciences
RESEARCH: Chemical Epigenetics, Immunoncology, Infectious Diseases, Metabolic Signalling, Targeted protein degradation
METHODS: Medicinal Chemistry, Synthesis, Assay development, Small Molecule Screening, Cellular target engagement analysis
TECHNOLOGIES: Organic synthesis infrastructure, Plate washer and readers, 400 MHz NMR for small molecule analysis
Jens Timmer 
Faculty of Mathematics and Physics – Institute of Physics
RESEARCH: Cancer Metabolism, Mathematical Modelling, Signal Transduction, Developmental Biology, Immune System, Synthetic Biology
METHODS: Dynamic Pathway Modelling, Parameter Estimation in Differential Equations, Uncertainty Analysis, Experimental Design
TECHNOLOGIES: High Performance Computing
Roland Schuele 
Faculty of Medicine and Medical Center – Department of Urology
RESEARCH: Epigenetic control of metabolic Signalling, Cancer Metabolism, Mitochondria, Immunometabolism, Reprogramming of tumor microenvironment, Immune Sensing, Epigenetics, Structural Biology, Cell Biology, Medicinal Epigenetics and drug development, Cancer
METHODS: Pharmacologic and Genetic Manipulation of Metabolism, Extracellular Metabolic Flux Analysis, Untargeted Metabolomics by MS and NMR (outsourced and via collaborations), 3D structure analyses, structure-based small molecule drug development
TECHNOLOGIES: Seahorse XFe96 Flux Analyzer (Agilent), Hypoxic Workstations i2 and H35 (Whitley), MS and NMR-based metabolomics, crystal structure analyses, medicinal chemistry
Anne-Kathrin Classen 
Faculty of Biology – Center for Biological Systems Analysis, CIAS
RESEARCH: Wound and Cancer Metabolism, Signal Transduction, Cell Biology, Developmental Biology, Chronic Wounds, Regeneration, Senescence
METHODS: Pharmacologic and Genetic Manipulation of Metabolism, Confocal Microscopy, Metabolome analysis (outsourced)
TECHNOLOGIES: In Vivo Drosophila Models, Intraepithelial and Inter-organ Communication
Robert Zeiser 
Faculty of Medicine and Medical Center – Internal Medicine I (Hematology and Oncology)
RESEARCH: Immunometabolism, Cancer Immunotherapy, Allogeneic Stem Cell Transplantation, Graft-versus-Host Disease, Oncogenic Signaling
METHODS: Pharmacologic and Genetic Manipulation of signaling in tumor cells, animal models for acute leukemia and melanoma, Metabolism, Extracellular Metabolic Flux Analysis, bioluminescence imaging
Felicitas Bucher 
Faculty of Medicine and Medical Center – Eye Center
RESEARCH: Signal Transduction, Metabolic Signalling, Mitochondria, Cell Biology, Angiogenesis, Retinal Vasoproliferative Disease
METHODS: Pharmacologic Manipulation of Metabolism, Extracellular Metabolic Flux Analysis (via collaboration), mouse models of retinal vasoproliferative disease (mouse model of oxygen-induced retinopathy, VLDLR Knockout mouse model, Laser-CNV model), RNAseq analysis (outsourced), untargeted Metabolomics (via collaboration)
Susana Andrade 
Faculty of Chemistry and Pharmacy – Institute for Biochemistry
RESEARCH: Metabolite signaling, Metabolite transport, Structure and function of Proteins, Channels, Transporters, Receptors, (Metallo-) Enzymes, Transport mechanisms, Receptor activation, Signal transduction, Protein-protein interaction
METHODS: Protein engineering and isolation, Affinity and size-exclusion chromatography, Molecular Biology, X-ray crystallography, cryo-EM, Proteoliposomes, Lipids, Detergents, Nanoparticles, SSM-/PLB-Electrophysiology, Microscale thermophoresis, Stopped-flow kinetics
TECHNOLOGIES: Planar Lipid Bilayer Electrophysiology (Warner Instr.), Solid-Supported Membrane Electrophysiology (Nanion Tech.), SX-20 Stopped Flow Spectrometer (Applied Photophysics)